Results¶
This section covers the results that are generated from a successful xiRT run. In the command line interface, the output folder needs to be specified. Typically, csv/xls files are the outputs of interest for most applications. The created folder will contain the following results:
- log file
- callbacks
- quality control visualizations
- tables (CSV/XLS)
For more details, please see the following paragraphs.
Log File¶
The log file contains useful information, including the xiRT version and parameters. Moreover the various steps performed during the analysis with xiRT are documented (e.g. number of duplicated entries, amino acid alphabet, maximum sequence length etc.). The logs also contain short numeric summaries from the CV training of xiRT.
2021-01-04 17:21:31,708 - xirt - INFO - Init logging file.
2021-01-04 17:21:31,708 - xirt - INFO - Starting Time: 17:21:31
2021-01-04 17:21:31,708 - xirt - INFO - Starting xiRT.
2021-01-04 17:21:31,708 - xirt - INFO - Using xiRT version: 1.0.63
2021-01-04 17:21:31,781 - xirt.__main__ - INFO - xi params: sample_data/xirt_params_3RT.yaml
2021-01-04 17:21:31,781 - xirt.__main__ - INFO - learning_params: sample_data/learning_params_training_cv.yaml
2021-01-04 17:21:31,781 - xirt.__main__ - INFO - peptides: sample_data/DSS_xisearch_fdr_CSM50percent.csv
2021-01-04 17:21:31,781 - xirt.predictor - INFO - Preprocessing peptides.
2021-01-04 17:21:31,781 - xirt.predictor - INFO - Input peptides: 17886
2021-01-04 17:21:31,781 - xirt.predictor - INFO - Reordering peptide sequences. (mode: crosslink)
2021-01-04 17:21:43,726 - xirt.processing - INFO - Preparing peptide sequences for columns: Peptide1,Peptide2
2021-01-04 17:21:44,296 - xirt.predictor - INFO - Duplicatad entries (by sequence only): 5426/17886
2021-01-04 17:21:44,312 - xirt.predictor - INFO - Encode crosslinked residues.
2021-01-04 17:21:46,910 - xirt.predictor - INFO - Applying length filter: 17886 peptides left
2021-01-04 17:21:46,920 - xirt.processing - INFO - Setting max_length to: 59
2021-01-04 17:21:47,012 - xirt.processing - INFO - alphabet: ['-OH' 'A' 'D' 'E' 'F' 'G' 'H' 'H-' 'I' 'K' 'L' 'M' 'N' 'O' 'P' 'Q' 'R'
'S' 'T' 'V' 'W' 'Y' 'clA' 'clD' 'clE' 'clF' 'clG' 'clI' 'clK' 'clL' 'clM'
'clN' 'clP' 'clQ' 'clR' 'clS' 'clT' 'clV' 'clY' 'clcmC' 'cloxM' 'cmC'
...
...
2021-01-04 17:28:38,903 - xirt.qc - INFO - Metrics: r2: 0.30 f1: 0.16 acc: 0.25 racc: 0.61
2021-01-04 17:28:39,207 - xirt.qc - INFO - QC: rp
2021-01-04 17:28:39,215 - xirt.qc - INFO - Metrics: r2: 0.69
2021-01-04 17:28:43,643 - xirt.__main__ - INFO - Writing output tables.
2021-01-04 17:29:01,207 - xirt.__main__ - INFO - Completed xiRT run.
2021-01-04 17:29:01,207 - xirt.__main__ - INFO - End Time: 17:29:01
2021-01-04 17:29:01,208 - xirt.__main__ - INFO - xiRT CV-training took: 7.20 minutes
2021-01-04 17:29:01,209 - xirt.__main__ - INFO - xiRT took: 7.49 minutes
Callbacks¶
Callbacks are used throughout xiRT to select the best performing model which is not necessarily the last (epoch) model trained. To reuse already trained models for transfer-learning and predictions on other data sets, the neural network model (“xirt_model_XX.h5”), as well as the parameters/weights (“xirt_weights_XX.h5”) are stored. In addition, training results per epoch are stored (“xirt_epochlog_XX.log”). “XX” refers to the cross-validation fold, e.g. 01, 02 and 03 for k=3; -1 refers the predictions for the ‘unvalidated’ fold (e.g. all PSMs/CSMS with FDR > e.g. 1%. The epoch log contains losses and metrics for the training and validation data. For some applications the used encoder (mapping of amino acids to integers) needs to be transferred. Therefore, the callbacks also include a trained label encoder from sklearn as pickled object (“encoder.p”). The last file additionally contains the formatted input data as pickled data. It can be used programmatically for debugging, exploration and manual retention time prediction using an already existing model. The data can be parsed in python via:
import pickle
X, y = pickle.load(open("Xy_data.p", "rb"))
alpha_peptides, beta_peptides = X[0], X[1]
# assuming 3 RT dimensions
RT1, RT2, RT3 = y
Visualizations¶
xiRT will create a rich set of QC plots that should always be investigated. The plots are stored in svg/pdf format.
Epoch Loss / Metrics¶
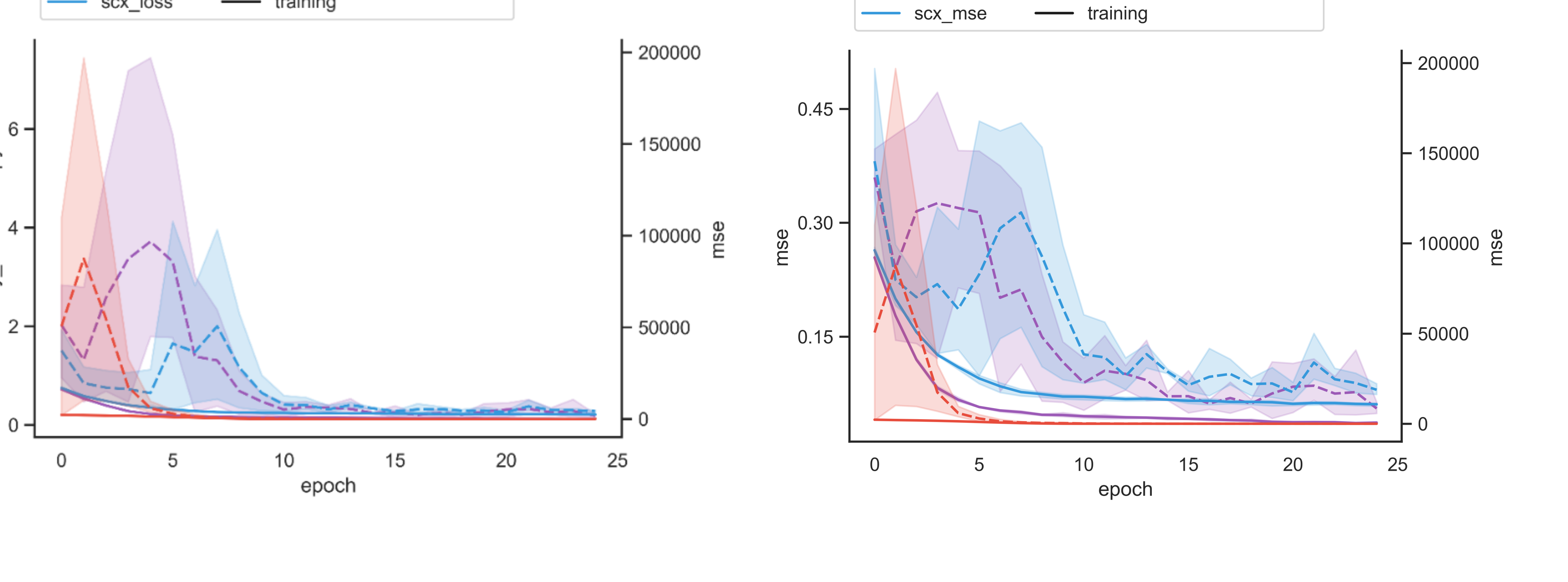
The epoch loss / metrics plot shows the training behavior over the epochs and is a good diagnostic tool to assess robustness across CV-folds, learning rate adjustment, overfit-detection and general learning behavior across tasks. In the example above, we see quick convergence and robust learning behavior after 10 epochs. In non-regression tasks, loss and metrics are not necessarily the same.
CV Summary¶
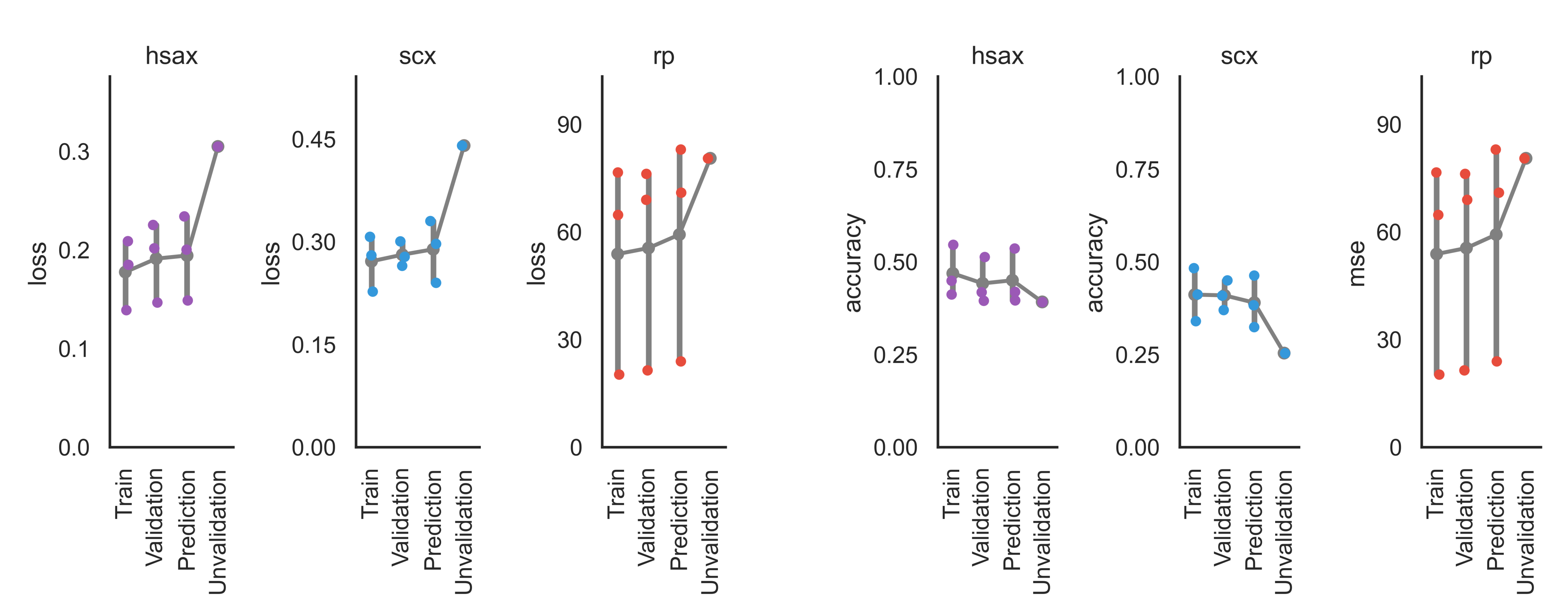
The CV summary shows the point estimates of the loss/metric for the training, validation and prediction folds for all training tasks. Unvalidation refers to the data not passing the training confidence (here: FDR) cutoff.
CV Observations¶
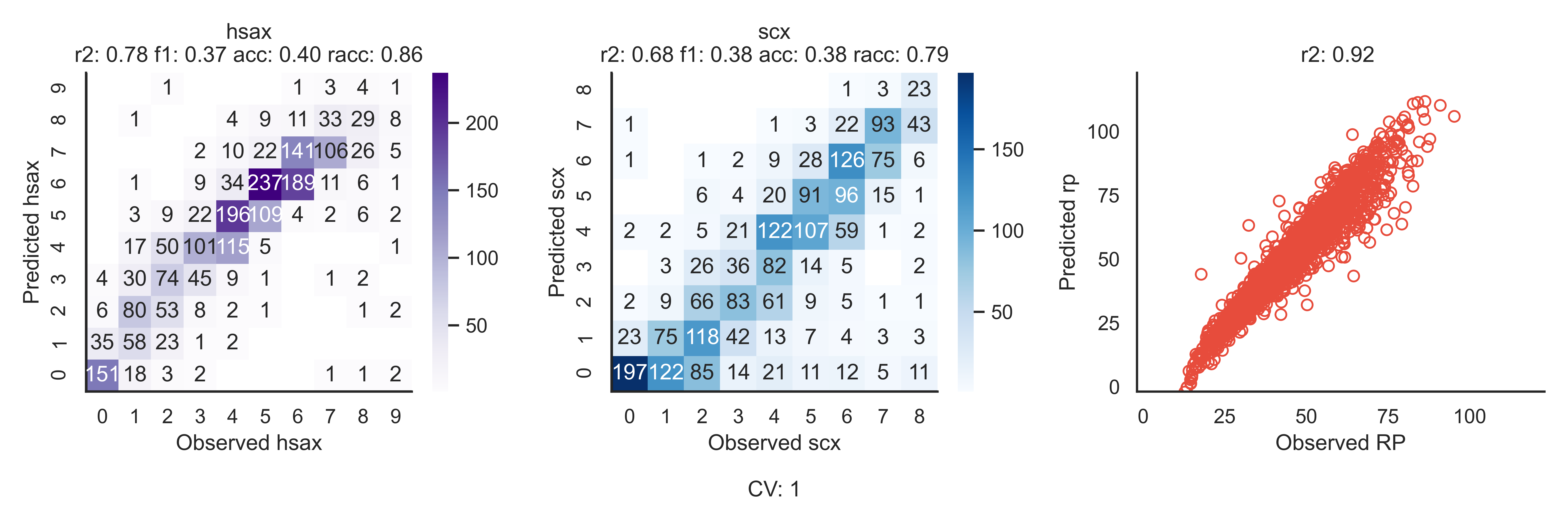
This plot shows the prediction performance for each CV-fold on all tasks. It also reports some key metrics that are not reported in the epoch log (r2, f1, accuracy, relaxed accuracy).
Tables¶
The tables contain a lot of extra information (some of which is used for the QC plots above). Please find an example of each file on (GitHub)[https://github.com/Rappsilber-Laboratory/xiRT/tree/master/sample_data/DSS_results_example].
Processed PSMs¶
This table (“processed_psms.csv”) contains the input data together with internally performed processing steps. The additional columns, as exemplified on a test dataset, are:
- swapped (indicator if peptide order was swapped)
- Seq_Peptide1/Seq_Peptide2 (peptide sequences in modX format)
- Seqar_Peptide1/Seqar_Peptide2 (peptide sequences in array format
- Duplicate (indicator if combination of sequences and charge is unique within the xiRT definition)
- scx0_based (0-based fraction number)
- scx_1hot (1-hot encoded fraction variable)
- scx_ordinal (ordinal encoded fraction variable)
- fdr_mask (indicator if PSM passed the FDR for training)
Epoch History¶
This table (“epoch_history.csv”) has similar data as the callbacks version but the CV results are concatenated and learning rate decay is documented.
Error Features¶
This table (“error_features.csv”) contains the input PSMID, crossvalidation split annotation and the predicted retention times (including their basic error terms).
Error Features Interactions¶
This table (“error_features_interactions.csv”) contains the input PSMID, and the some engineered error terms from the previous table.
Model Summary¶
This table (“model_summary.csv”) contains important metrics that summarize the performance of the learned models across CV-splits and their corresponding train/validation/prediction splits.